Proteins help genes do their job of creating proteins in an endless cycle of creation and control that is essential to life
We often hear about DNA, the double-helix made up of nucleic-acid that makes life as we know it possible. We hear about how damage to DNA can cause disease1 and cause our cells—the most basic unit of life—to die2. We hear a lot about how important DNA is, about how it makes up our genetic code and how critical it is to keep that genetic code healthy and safe. But what is our DNA actually doing for us?
When we boil it down to the basics, the sole purpose of our DNA, with extremely rare exception3, is to create proteins. Proteins are large, complex molecules that are the real workers of our cells; they help give our cells structure, serve as transport molecules, work as antibodies to fight infection, and facilitate important biochemical reactions4. Without proteins, we wouldn’t have much control over how our cells function and respond to the world around us, and this control is essential to survival.
Genes are blueprints written in DNA, and the genome is a blueprint library for proteins
The cellular machinery of genes and proteins
Think of our cells as an automated factory. Our DNA is a blueprint library, where all the information for all the different products is safely stored. A computer decides—from various inputs and signals—which product must be made, and it selects and unravels the correct blueprint for the situation. Instead of risking damage to the original copy of a blueprint out on the factory floor, another machine copies it word for word, line for line, with remarkable accuracy. The resulting transcript is sent out from the blueprint room to the factory floor. There, the transcript is placed into a machine that reads it and assembles the desired product. In our cells, the transcript is called messenger RNA, the assembly machine is called a ribosome, and the many different possible products are proteins.
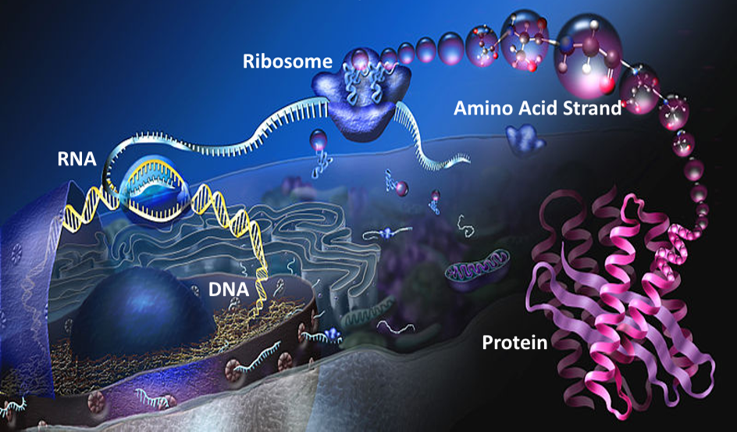
DNA in the nucleus is ‘read’ and copied into mRNA, then ribosomes produce an amino acid strand that folds into a functional protein.
Source: Modified from How Proteins are Made, Nicolle Rager, National Science Foundation
It gets a bit more complicated than just that, however. That’s because at each step of this production line, changes can be made to the information retrieved from the blueprint library. One product can be chosen for production along with, instead of, or in greater or lesser abundance than another; this reflects how our cells choose when to express different genes—when to turn DNA into protein or not.
There are countless ways for the cell to control what product is produced despite using the same original blueprint.
The factory system can also make direct edits to the transcript of the blueprint, it can literally change the product’s design by modifying the transcript after it’s been made but before it gets to the assembly machine5. It can destroy the transcript before it’s translated into product6 or block the assembly machine from ever reading the transcript7, both are ways to prevent the final product from being produced. This cellular factory can even change the accuracy with which the assembly machine reads the transcript8 and make modifications to the products after they’ve been made9. There are countless ways for the cell to control what product is produced despite using the same original blueprint. In humans, these mechanisms greatly increase our control over how our genome is expressed. They increase the number of protein variations we can produce from approximately 20,000 that are encoded directly in our DNA to multiple millions of forms10.
Proteins have an essential role in helping genes create other proteins
Many of the products this automated factory produces are for running the factory itself. That computer from before, the one that decides which blueprint to turn into a product, is made up of parts produced by its own factory. All of the components involved in altering and controlling the blueprints in transit and even the assembly machines themselves are made in-house. These factory machines work in tandem to build new versions of themselves, with countless variations, by using and manipulating the information retrieved from the blueprint library.
Genes and proteins exist on a two-way road, not a one-way street—proteins help genes to create proteins in an endless cycle of creation and control that is essential to life.
Our cells do this. Proteins control cell structure, cellular component transport, and chemical reactions all throughout the cell, including those parts involved in this production process. They help control which genes are turned into proteins and how those genes are altered during production11. Moreover, when DNA is damaged, proteins are what rush to the scene to repair our genetic code12. Genes and proteins exist on a two-way road, not a one-way street—proteins help genes to create proteins in an endless cycle of creation and control that is essential to life.
Why your proteome is vulnerable to downward spirals
The issue with cycles is that they can turn into spirals. Since our DNA is so important in maintaining our proteins, certain damage to our genome can impair parts of our protein population (known, like genome is to genes, as the proteome). If that specific part of our proteome is essential in DNA regulation—for example, in repairing damage to DNA—the two-way road of DNA-protein interdependence can turn into an uncontrolled tailspin towards the development of disease1 and cell death2. Likewise, something that damages our DNA-repair-proteins directly may make our genome more vulnerable to damage. Damage to any other set of proteins, at the very least, reduces the genome’s control over that protein’s area of function—if the final product our genome is trying to make isn’t working, that aspect of our genome is temporarily useless. This damage to proteins can also lead to any number of biological errors that further reduce the health of our cells. The result is a downward spiral.
The role of oxidative damage
Oxidative damage is a perfect example of a wrench thrown into the cog of our genome-to-proteome assembly line.
Unfortunately, there are many ways for damage to DNA and proteins to occur, sometimes both types of molecules are damaged at once. One of the most important sources of damage in the fields of biomedicine and human health is oxidative stress, which is often just called oxidative damage. Oxidative damage occurs when reactive molecules known as reactive oxygen species (ROS) accumulate past our cells’ capacity to control them13 and begin to damage vital cell components including proteins and DNA14–17. While ROS are a normal part of our biology, in high enough abundance they can lead to cell death18, and there are many different ways that ROS-associated oxidative damage can be brought on, ranging from our own immune responses to exercise8,14,19–26.
Oxidative damage is a perfect example of a wrench thrown into the cog of our genome-to-proteome assembly line. Naturally, our bodies have defense mechanisms at the ready. To understand these defenses and how to boost them, it is critical to understand that proteins are three-dimensional molecules that must fold into their correct shape in order to function properly27. When a protein is attacked by ROS, it undergoes chemical changes that can disrupt the stability of its structure and cause it to unfold, losing its function28. This is why, in response to oxidative stress, our body not only produces antioxidant molecules to lower ROS levels directly, but also mobilizes a response to help proteins refold24,29,30 so that our cells regain their health and production efficiency.
How to support your proteome and address oxidative damage
It is here that Eng3’s NanoVi device steps into the mix. Using specific wavelengths of electromagnetic energy, NanoVi is able to enhance the formation of something called ordered or exclusion-zone (EZ) water throughout the body. Ordered water possesses a structural quality that has been observed to greatly increase protein stability and the rate of protein folding31. This increase in protein folding efficiency leads to more efficient cellular production in general and specifically applies to our example of oxidative stress causing damage to proteins and DNA.
By bringing a naturally-driven yet powerful ability to increase the health and efficiency of our proteome, NanoVi promotes the well-being of the entire genome-to-proteome production line.
By boosting protein stability, NanoVi helps to protect proteins from unfolding as a result of an oxidative attack. Likely related to this ability to keep proteins (such as DNA-repair proteins) functioning, research performed at the IMSB Olympic Training Centre in Vienna, Austria shows significantly reduced DNA damage in athletes given NanoVi sessions along with their exercise regimens32. The levels of ROS-neutralizing antioxidant proteins in humans are also significantly increased by NanoVi, and even those proteins that are damaged by oxidation recover significantly faster33 when exposed to NanoVi.
With this evidence at hand, it is easy to see how NanoVi technology fits perfectly into the world of genetics and genetic regulation. Genes are essential to making proteins, yes, but proteins are essential to all our cells functions—including the integrity of our DNA. By bringing a naturally-driven yet powerful ability to increase the health and efficiency of our proteome, NanoVi promotes the well-being of the entire genome-to-proteome production line. This boost, in turn, is what makes NanoVi such a particularly effective tool for improving health, performance, recovery, and healing—all beginning at the molecular level.
If you’d like to learn more about the NanoVi device, its proven benefits, and the price, sign in below.
Eng3 will send you an email as soon as you submit the form, please check your spam filter if you don’t receive it. We only send study results and other valuable information. Your email will not be shared or flooded with promotional content.
References
1. Zhang, W., van Gent, D. C., Incrocci, L., van Weerden, W. M. & Nonnekens, J. Role of the DNA damage response in prostate cancer formation, progression and treatment. Prostate Cancer Prostatic Dis. (2019). doi:10.1038/s41391-019-0153-2
2. Surova, O. & Zhivotovsky, B. Various modes of cell death induced by DNA damage. Oncogene 32, 3789–3797 (2013).
3. Papayannopoulos, V. Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 18, 134–147 (2018).
4. B, A., A, J. & J, L. Protein Function. in Molecular Biology of the Cell (Garland Science, 2002).
6. Gebert, L. F. R. & MacRae, I. J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20, 21–37 (2019).
7. Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009).
8. Netzer, N. et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462, 522–526 (2009).
9. Biggar, K. K. & Li, S. S.-C. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 16, 5–17 (2015).
10. Minde, D.-P., Ramakrishna, M. & Lilley, K. S. Biotin proximity tagging favours unfolded proteins and enables the study of intrinsically disordered regions. Commun. Biol. 3, 38 (2020).
11. Arrowsmith, C. H., Bountra, C., Fish, P. V, Lee, K. & Schapira, M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 (2012).
12. Scully, R., Panday, A., Elango, R. & Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 (2019).
13. Pizzino, G. et al. Oxidative Stress: Harms and Benefits for Human Health. Oxid. Med. Cell. Longev. 2017, 8416763 (2017).
14. Schieber, M. & Chandel, N. S. ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462 (2014).
15. Lobo, V., Patil, A., Phatak, A. & Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 4, 118–126 (2010).
16. Phaniendra, A., Jestadi, D. B. & Periyasamy, L. Free radicals: properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 30, 11–26 (2015).
17. Cadet, J. & Wagner, J. R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 5, a012559 (2013).
18. Ghosh, N., Das, A., Chaffee, S., Roy, S. & Sen, C. K. Reactive Oxygen Species, Oxidative Damage and Cell Death. in Immunity and Inflammation in Health and Disease: Emerging Roles of Nutraceuticals and Functional Foods in Immune Support (eds. Chatterjee, S., Jungraithmayr, W. & Bagchi, D. B. T.-I. and I. in H. and D.) 45–55 (Academic Press, 2018). doi:https://doi.org/10.1016/B978-0-12-805417-8.00004-4
19. Watts, M. E., Pocock, R. & Claudianos, C. Brain Energy and Oxygen Metabolism: Emerging Role in Normal Function and Disease . Frontiers in Molecular Neuroscience 11, 216 (2018).
20. Sanders, S. P. et al. Hyperoxic sheep pulmonary microvascular endothelial cells generate free radicals via mitochondrial electron transport. J. Clin. Invest. 91, 46–52 (1993).
21. Haffor, A. S. . Effects of O2 Breathing on Cardiac Mitochondrial, GOT and Free Radical Production. J. Med. Sci. 4, 164–169 (2004).
22. Alttas, O. & Haffor, A.-S. Effects of hyperoxia periodic training on free radicals production, biological antioxidants potential and lactate dehydrogenase activity in the lungs of rats, Rattus norvigicus. Saudi J. Biol. Sci. 17, 65–71 (2010).
23. Lushchak, V. I. et al. Hyperoxia results in transient oxidative stress and an adaptive response by antioxidant enzymes in goldfish tissues. Int. J. Biochem. Cell Biol. 37, 1670–1680 (2005).
24. Dimauro, I., Mercatelli, N. & Caporossi, D. Exercise-induced ROS in heat shock proteins response. Free Radic. Biol. Med. 98, 46–55 (2016).
25. Alessio, H. M. Exercise-induced oxidative stress. Med. Sci. Sports Exerc. 25, 218—224 (1993).
26. Powers, S. K. & Jackson, M. J. Exercise-Induced Oxidative Stress: Cellular Mechanisms and Impact on Muscle Force Production. Physiol. Rev. 88, 1243–1276 (2008).
27. Dobson, C. M. Protein folding and misfolding. Nature 426, 884–890 (2003).
28. Di Domenico, F., Head, E., Butterfield, D. A. & Perluigi, M. Oxidative Stress and Proteostasis Network: Culprit and Casualty of Alzheimer’s-Like Neurodegeneration. Adv. Geriatr. 2014, 527518 (2014).
29. Zhang, Z. et al. Redox signaling and unfolded protein response coordinate cell fate decisions under ER stress. Redox Biol. 25, 101047 (2019).
30. Holtz, W. A., Turetzky, J. M., Jong, Y.-J. I. & O’Malley, K. L. Oxidative stress-triggered unfolded protein response is upstream of intrinsic cell death evoked by parkinsonian mimetics. J. Neurochem. 99, 54–69 (2006).
31. Zuo, G., Hu, J. & Fang, H. Effect of the ordered water on protein folding: An off-lattice Go-like model study. Phys. Rev. E 79, 31925 (2009).
32. Dörr, W. & Bozsaky, E. Improved Regeneration of DNA Double Strand Breaks with NanoVi(TM) Bio-identical Signaling Technology. (2014).
33. Yablonskaya, O. I., Trofimov, A. V., Voeikov, V. L., Novikov, K. N. & Buravleva, E. V. Evidence of Protective and Regenerating Properties of NanoVi water vapor. in Annual Water Conference 1 (2018).




