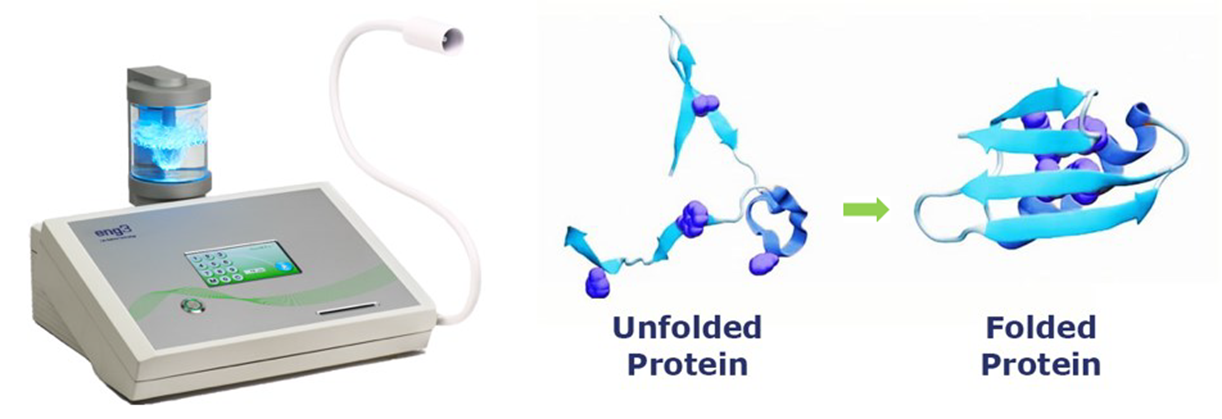
The protein folding problem that challenged scientists for 50 years has succumbed to the technical advances of 2021
Excitement about protein folding spilled into the public eye in late 2018. DeepMind’s artificial intelligence program, called AlphaFold, demonstrated the ability to predict folded protein structures from their chains of amino acids with about 75% accuracy. At that time DeepMind’s CEO stated: “We’ve not solved the protein folding problem, this is just a first step. It’s a hugely challenging problem, but we have a good system and we have a ton of ideas we haven’t implemented yet”.
Now another big step has been taken. Big enough to win Science’s award for the scientific breakthrough of 2021. Below is a clip announcing award winners. If you want to skip to first place and the prediction of the shape of folded proteins, jump to 3:35 in the video.
AlphaFold & AlphaFold 2
Scientific breakthrough of the year awarded by Science,
the American Associationfor the Advancement of Science (AAAS)
Scientific breakthrough of the year awarded by Science,
the American Associationfor the Advancement of Science (AAAS)
This breakthrough in predicting the structures of proteins is a double win, according to Science Editor-in-Chief Holden Thorp:
“First, it solves a scientific problem that has been on the to-do list for 50 years… Second, it’s a game-changing technique that, like CRISPR or cryo-EM, will greatly accelerate scientific discovery.”- Holden Thorp, Science Editor-in-Chief
To put Dr. Thorp’s comment in context, the scientists behind CRISPR and cryo-EM earned Nobel Prizes.
Who won? AlphaFold and RoseTTAFold
Although the video does not get into RoseTTAFold, Science’s award acknowledges both DeepMind and the University of Washington’s Institute for Protein Design (IPD) for developing RoseTTAFold. IPD’s significant contribution reduced the processing time needed to predict the folded shape of a protein. Prior to RoseTTAFold, protein prediction models took a great deal of time because the modeling is so complex. IPD made it possible to predict a protein structure in as little as 10 minutes.

Scientific innovations of this magnitude are made possible by collaboration. IPD and DeepMind have made their code open source, which accelerates protein research.
Both Google’s DeepMind and IPD at the University of Washington have made their technology open source. IPD’s collaboration with other research teams in Canada and Great Britain contributed to their remarkable improvement in computation times. A collaborative approach and open-source code have contributed to the incredible speed with which hundreds of thousands of protein structures have been modeled in 2021 alone.
When computer modeling and the use of artificial intelligence were applied to the protein-folding problem, previous methods could be used to verify the results. Here is a taste of the path to deciphering the 3-D shape of proteins.
A history rich in Nobel Laureates
In 1957, scientists at Cambridge used X-ray crystallography to determine the first protein structures. This earned them the 1962 Nobel Prize in Chemistry. Shortly after this, three American Biochemists established the connection between the sequence of amino acids that make up a protein and its folded shape. This earned them the 1972 Nobel Prize in Chemistry. Once it was understood that the sequence of amino acids could predict the three-dimensional structure of a protein, figuring out the shape became known as the “protein-folding problem”. This problem has challenged scientists for the last half century.
The X-ray crystallography prediction method, although slow and labor-intensive, was a reliable way to reveal the folded shape of proteins. The development of cryo–electron microscopy (cryo-EM) made things much faster. Using very expensive equipment, cryo-EM could determine protein structures without using crystalized samples. Swiss scientists added Nuclear magnetic resonance spectroscopy of proteins (protein NMR) to the mix. Protein NMR provided information about both the structure and dynamics of proteins. Yet another Nobel Prize in Chemistry (1991) was awarded for this contribution.
Why the “protein folding problem” is so important
Knowing the folded shapes of proteins has been central to medical research for decades. Most therapeutic drugs currently on the market are based in proteomics. What has changed is that pharmaceutical companies no longer have to do it the hard way, using the expensive and time-consuming methods outlined above. The 2021 breakthrough in protein structure prediction help researchers develop interventions faster and better understand the interaction between proteins. AI protein folding modeling also offers the potential to design new proteins, ones that don’t exist in nature. These artificial proteins could be used for special purposes including individualized therapies and specialized materials.
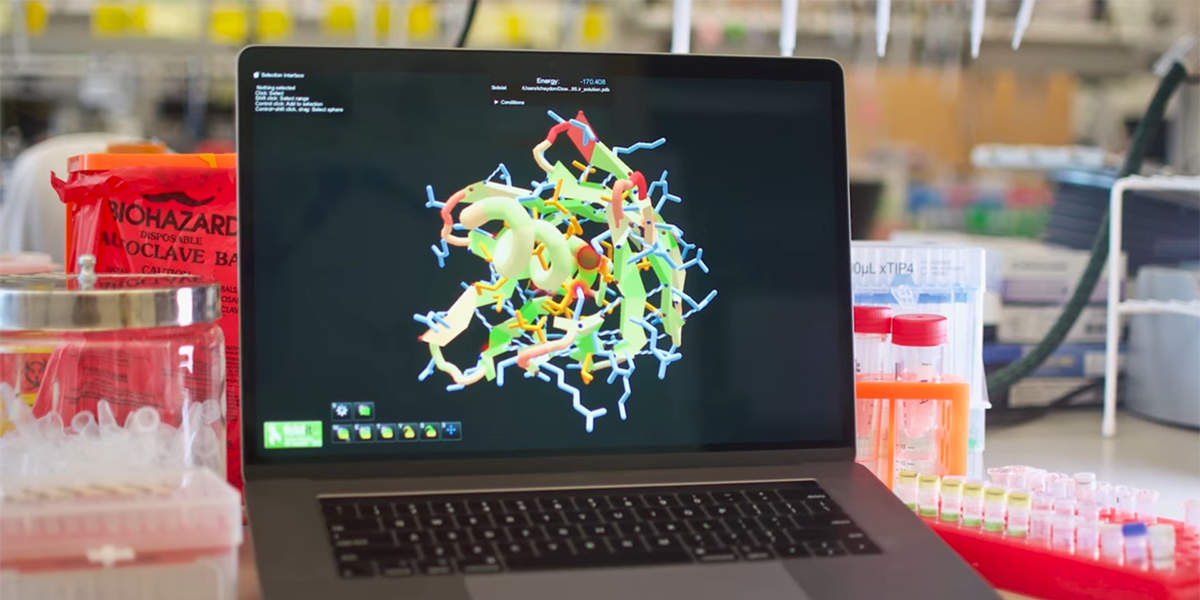
AI driven modeling of protein structures enables researchers to develop interventions faster and at lower cost than previous methods.
“Because proteins are complicated in structure and the protein folding problem hadn’t really been well understood until recently, the approach was to take naturally occurring proteins and modify them by introducing random mutations and selecting for the most useful variants to solve specific problems… As we get closer to solving the protein folding problem, we can now design completely new proteins from scratch for specific purposes.”
– David Baker, Director of UW Institute of Protein Design
Proteins cannot function until they are folded, and their final shape is encoded in the protein itself. In addition, the work a protein does in the body is determined only by its folded shape. The importance of protein folding cannot be overstated since all of life requires this final step in the creation of working proteins. Supporting protein folding is the whole point of NanoVi® technology.
NanoVi’s unique approach is to trigger protein folding by influencing the water that surrounds all proteins. By improving the environment, it is not necessary to determine a protein’s folded shape to help it fold more readily. NanoVi supports all proteins, whether their final folded shape is known or not.
If you’d like to learn more about the NanoVi device, its proven benefits, and the price, sign in below.
Eng3 will send you an email as soon as you submit the form, please check your spam filter if you don’t receive it. We only send study results and other valuable information. Your email will not be shared or flooded with promotional content.
Reference Articles
1. Wiggers, K. Deepmind’s AlphaFold wins CASP13 protein-folding competition. The Machine https://venturebeat.com/2018/12/03/deepminds-alphafold-wins-casp13-protein-folding-competition/ (2018).
2. Beckwith, W. Science’s 2021 Breakthrough: AI-powered Protein Prediction | American Association for the Advancement of Science. AAAS https://www.aaas.org/news/sciences-2021-breakthrough-ai-powered-protein-prediction (2021).
3. Holden, T. H. Proteins, proteins everywhere. Science 374, 1415 (2021).
4. Schubert, C. Univ. of Washington AI protein folding discovery wins ‘Breakthrough of the Year’ award from Science. GeekWire (2021).
5. Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
6. Nair, P. QnAs with David Baker. Proceedings of the National Academy of Sciences 115, 13142–13144 (2018).
Video Transcript for Breakthrough of the Year 2021 – first prize
For more than 50 years, scientists have tried to understand how proteins, the building blocks of biology, transform from simple chains of amino acids into precisely folded 3-D structures. Those structures determine how proteins interact with other molecules and could reveal everything from unknown drug targets to new insights into basic biology. This year the power of artificial intelligence improved those predictions by magnitudes.
Before AI entered the scene, scientists used experimental techniques to map proteins atom by atom and researchers have used these methods to uncover the structures of more than 180,000 proteins. But such work is slow and expensive. To speed things up, scientists started using computers to predict the shapes that proteins might take. For the last two decades those predictions have gotten better but they still have room for improvement. Then AI changed everything.
AlphaFold, a program developed by Google’s sister company DeepMind, outshone every team in a major competition in 2018, and in 2020 AlphaFold2 showed that it could predict proteins as well as experimental techniques, a major milestone. And this year researchers solved 350,000 human protein structures, 44% of all known human proteins and a really good fraction of the 200,000,000 thought to exist. Other researchers are using AI to predict how proteins come together and interact for a wide variety of applications. Even now AlphaFold2 is modeling the spike protein in the new Omicron variant of SARS-CoV-2 and scientists are eagerly awaiting results.
rev01